Hepatitis B Virus Dna Detectable Again
Introduction
Occult hepatitis B virus (HBV) infection (OBI) is characterized by the presence of very low levels of HBV DNA in the plasma and/or in the liver, with undetectable hepatitis B surface antigen (HBsAg), with or without antibodies to hepatitis B core antigen (anti-HBc) or hepatitis B surface antibody (anti-HBs), outside the pre-seroconversion window period (1), and OBIs may contribute to the exacerbation of acute HBV infection and the development of HBV-associated cirrhosis and hepatocellular carcinoma (HCC) (2). OBI has been found in blood donors, and the prevalence varies from less than 1 to 16% dependent on the endemicity of HBV infection, the screening NAT assays, and confirmation algorithms used (iii–6). Although donations with OBI pose a significant risk of transfusion-transmitted HBV infection, OBI screening is very difficult due to intermittent very low virus load and mutations in HBV genome (7, 8).
The blood screening strategy past employing both HBsAg and anti-HBc detection combined with HBV NAT assay in some developed countries allows the detection of window-menstruation infection, OBI, and HBV mutated strains possible to maximally ensure blood safety. Nevertheless, in HBV owned countries such equally in Cathay, deferral of all anti-HBc positive donations was impractical due to the shortage of claret supply. In addition, high cost of ID-NAT prompt users to adopt screening strategies of pooling of multiple donor samples (nine). As a pilot NAT project initiated by the National Wellness Commission (NHC) of China, Shenzhen Blood Heart has been using the Roche Cobas TaqScreen MPX test2.0 for HBV DNA, hepatitis C virus (HCV) RNA, and human immune-deficiency virus (HIV) RNA in routine screening in MP6 (pooled from vi individual donations) format since 2017. It turns out this assay is highly specific, sensitive, reproducible, and robust (ten, 11). However, unlike proportions of MP-reactive but private not-reactive donations (MP+/ID−) take been observed during daily screening. In ID-NAT screening, non-repeat-reactive (NRR) donations are not released for transfusion in near countries. However, for MP6-NAT it is non an option to discard the units implicated in non-resolved pools, leaving a fatal threat to claret rubber, particularly in HBV loftier prevalent countries such as China.
Previously, we take reported nearly half of the initial reactive, but further discriminatory test negative donations were identified as OBIs, which strongly suggested that a proportion of donations with ID-NAT screening NRR results might pose HBV manual risk fifty-fifty when using highly sensitive NAT assays (12). In this study, we move on to further analyze the not-resolved reactive mini pools in respect to their potential transmission risk for HBV infection. In society to clarify and evaluate the true infection status of the non-resolved donations, alternative Ultrio Plus ID-NAT, nested PCR for S, BCP/PC, and qPCR with high volume extraction were performed (13), and HBV infection status volition be characterized from these non-resolved donations. The optimized screening strategy by adding anti-HBc testing volition also be discussed.
Materials and Methods
Subjects and Samples
As a routine exercise, all donations were collected and screened serologically as reported in our previous study (12). In total, 103,356 seronegative blood samples (17,226 MP6 pools) were enrolled in this written report for further analysis.
NAT Screening of Donor Samples
Six private donated blood were pooled into 1 mini puddle (MP6), and NAT screening of the MP6 was performed on a fully automated Roche Cobas due south 201 system using a multiplex polymerase concatenation reaction kit (Cobas TaqScreen MPX test, version 2.0, Roche Molecular Systems, Branchburg, NJ, The states; 1 ml, LOD: 2.iii IU/ml for HBV Deoxyribonucleic acid, i IU/ml=5.vi copies/ml). Donations with negative NAT results were released, and MPX-reactive MP6 were resolved past retesting each private donation (ID-NAT). Samples individually reactive were classified as MPX echo reactive (designated every bit NAT+). If ane NAT+ was identified, other v non-reactive donations were designated as NAT− and tin can be released. If all 6 private donations in the pool were detected as MPX non-reactive, they were classified equally MP non-resolved donations. These non-resolved donations were farther tested individually by an alternative Procleix Ultrio Plus HBV discriminatory assay (Grifols Diagnostic Solutions, Inc. and Hologic; 0.5 ml, LOD: 3.4 IU/ml) to identify the presence of HBV DNA. Donations showing not-reactivity in the Ultrio Plus assay were classified as NAT non-resolved donations by Ultrio Plus MPX. Donations showing reactivity in discriminatory HBV assays were identified as HBV NAT+, and the pool was regarded equally resolved pool by Ultrio Plus.
Serological and Molecular Detection
All resolved and non-resolved samples, including frozen plasma, were nerveless for farther decision. HBsAg, anti-HBs, HBeAg, anti-HBe, and anti-HBc were tested by commercially available electrochemiluminescence immunoassay, ECLI (Roche, Usa). All samples were retested on anti-HBc past a domestic Eia kits (WanTai Diagnostics, Beijing, China), and only samples with reactivity for both assays were designated as anti-HBc+.
HBV Deoxyribonucleic acid was additionally extracted from 2.5 ml of plasma by HighPure Viral Nucleic Acid Large Book Kits (Roche Diagnostics Gmbh, Mannhein, Germany) and were analyzed past a combination of qPCR (13) (LOD: 5 IU/ml) and nested PCRs (the LOD for the combination qPCR/nested PCR assay used with high book extraction can reach as low as i IU/ml (5.6 copies/ml)), which amplified BCP/PC (LOD: ten IU/ml) and S regions (LOD: 10 IU/ml) as previously described (4, fourteen).
HBV Dna Sequencing and Genotyping
The amplified products of BCP/PC and S regions were sent to Shanghai Invitrogen Co., Ltd. (Guangzhou, Cathay) for sequencing. HBV genotyping was performed past Phylogenetic analysis using MEGA7.1 program, and the neighbor-joining method based on Kimura 2-parameter mode and complete deletion for gaps with 1,000 bootstrap replications was chosen every bit previously reported (13). So-chosen wild blazon consensus sequences were derived from the alignment of 124 genotype B and 95 genotype C sequences from HBsAg+ blood donors (iv).
Statistical Analyses
Stata 16.0 was applied for statistical assay of diverse data. P < 0.05 (ii-tailed) was set equally the meaning cut-off level.
Results
Screening Results of the Blood Donations
A total of 103,356 seronegative blood donations were nerveless. A total of 17,226 pools were derived, and 98 pools were reactive for HBV DNA by MPX2.0 NAT in MP6 format. Fifty-six pools (56/98, 57.i%) were resolved with HBV DNA+ (MP+/ID+), and 42 pools (42.9%, 252 donations) were detected non-reactive past MPX2.0 ID-NAT (MP+/ID−, Effigy 1), including 169 males and 83 females (Table one). The kickoff-time and repeat donors were 115 (45.6%) and 137 (54.iv%), respectively.
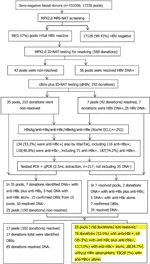
Effigy 1 Menses chart of serological and molecular identification of MPX ii.0 MP6-NAT in non-resolved samples.

Table 1 Demographic and viral characteristics of 252 (42 pools) not-resolved blood donations.
Supplemental Serological Testing Results for 252 Non-resolved Donations
After tested by Elecsys II assay for HBsAg, anti-HBs, HBeAg, anti-HBe, and anti-HBc, 134 out of the 252 non-resolved donations (53.2%) were reactive for anti-HBc, all of which were confirmed positive by WanTai anti-HBc kit. In that location was no correlation between the presence of anti-HBc and gender or donor type. In contrast, in that location was a clear increase of anti-HBc prevalence with age, ranging from 32.3% in the <thirty age group to 79.ii% in the >50 age group (χ2 = 34.2, p=0.00).
Ultrio Plus ID-NAT, Nested PCR, and qPCR Testing for HBV Dna of Non-Resolved Donations
HBV Deoxyribonucleic acid was further analyzed individually for the 252 not-resolved donations past Ultrio plus dHBV, and seven donations out of 42 MPs (252 donations) (7/252 = ii.8%) were resolved as dHBV+, leaving 210 donations notwithstanding non-resolved. After the 210 non-resolved samples and 7 dHBV+ samples were farther analyzed by nested PCR (BCP/PC and S region) and qPCR with 2.five ml extraction of Deoxyribonucleic acid individually, 10 donations out of 35 MPs (150 donations) were identified as HBV DNA+ (Table 2). All together, 17 donations from 42 MPs (252 donations) were resolved HBV Dna+ (one per pool).

Table 2 The serological and molecular label results of the 17 identified HBV Dna+ donations.
Of the 17 identified HBV Deoxyribonucleic acid positive donors, seven were repeat donors and x were start-time donors, and viii were males and nine were females with boilerplate age of 36.2. Regarding the other viral markers, viii had anti-HBc solitary, one carried anti-HBc and anti-HBe, and eight were positive for both anti-HBc and anti-HBs. Three out of the eight anti-HBs positive samples had titers over 100 IU/50. Finally, 150 donations still remained non-resolved, among which 17 were identified equally OBIs, and 85 donations were resolved HBV Deoxyribonucleic acid negative.
Comparison of Seromarkers Distribution Among Not-Resolved 252 Donations, Remaining 150 Non-Resolved Donations, and Resolved Negative Donations
In the remaining non-resolved donations, 79/150 (52.6%) donations carried anti-HBc, in which xi cases (7.3%) were anti-HBc alone. While, in the resolved negative donations from 17 resolved pools (85 donations), 38/85 (44.7%) donations carried anti-HBc, including 1 (one.two%) with anti-HBc alone (Tabular array three). The rate of anti-HBc alone in the remaining non-resolved donations is half dozen times higher than that in the resolved negative donations (P<0.05).

Table 3 HBV seromarker distribution of non-resolved donations, the remaining non-resolved and resolved negative donations.
Genotyping and Mutation Analysis of the Southward and BCP/PC Regions in the Identified HBV DNA+ Donations
Totally, 17 samples (17/42, twoscore.5%) in the non-resolved pools were identified HBV Dna+ and classified as OBIs by Ultrio plus and additionally serological and molecular assays. The maximum and median viral loads were 37.two IU/ml and 5.1 IU/ml, respectively. The phylogenetic analysis identified 7 donations with genotype B and 1 genotype C and ane genotype D. The South region amino acrid sequences of these nine cases showed that all cases had amino acrid substitutions. Regarding the seven genotype B samples, two out of 7 (28.6%) samples were observed equally wild type (L017 and Q84). Iv samples (57.1%) had vaccine escape mutations: G112R (Q8), T126S (L012), M133T (Q27), D144E (Q139), and S174N (Q139). Sample Q27 with anti-HBs harbored a T131N/M133T Northward-glycosylation mutation, which may interfere with recognition of HBsAg by anti-HBs, therefore contributing to virus escape from the host immune organization (15). In addition, four/7 (57.1%) accept mutations that have potential affect on the detection of HBsAg: T126S (L012), M133T (Q27), F134L (Q27), T143L (Q8), L175S (Q139). Various mutations associated with OBIs—Q101R (N022 and Q139), P105R (Q139), T126S (L012), P127H (Q27), M133T (Q27), S174N (Q139), and V177A (Q139)—were besides detected in four (57.1%) samples. For the 1 genotype C sequence, information technology harbored multiple mutations including Q30K, L53S, T113K, and T118K, and lastly a genotype D sample conveying V96A, L104W, G112E, P127L, Q129P, and S164N mutations.
BCP/PC genes were amplified from six samples by nested PCR. Nucleotide mutations with loftier frequency were found as follows: 5/six (83.3%) sequences contain T1719G, which has been previously reported to inhibit HBV replication through Enh II and HBx proteins mutation in vitro (xvi). Likewise present were 6/6 (100%) A1726C and 5/6 (83.3%) C1730G mutations. In addition, other mutations such as A1752G/T (33.iii%), C1773T (33.3%), G1809T (33.iii%), A1846T (33.3%), G1799C (xvi.7%), C1853T (16.vii%), G1896A (16.7%), G1899A (16.7%), and G1915A (16.seven%) were as well observed.
Discussion
Donor screening is essential to ensure blood prophylactic to forestall transfusion-transmitted infections. To decrease the remainder take chances for HBV infection, both HBsAg and anti-HBc, together with highly sensitive HBV NAT (ideally ID-NAT) screening, provide the highest level of blood safety for recipients, merely this comprehensive screening strategy is simply adopted in high income countries due to high cost and strict requirement of NAT (17). In most developing countries, especially China with a high positive rate for anti-HBc, alternative approaches tin be used to remainder the cost and safe. Currently HBsAg and MP-NAT screening accept been used in China to prevent near cases of HBV infections from transfusion. In countries where NAT screening is not implemented, screening HBsAg and anti-HBc could place donors with OBI at relatively low toll. Then, testing donations with HBsAg negative but anti-HBc positive using a highly sensitive NAT for HBV-Deoxyribonucleic acid could intercept nigh acute HBV infections within window period (WP) or OBIs. These two unlike rubber procedures applied in sequence could guarantee claret safety at a relatively low toll. As a compromised strategy, most Chinese claret centers adopt both HBsAg and HBV MP-NAT screening to balance the cost and safe by shortening the WP and intercepting most OBIs. The same approach has also been applied in Taiwan (18). In a country with a high positive rate for anti-HBc, the use of anti-HBc positive/HBsAg negative/HBV-Dna negative donations for anti-HBc (>100 UI) and/or HBsAg positive individuals should also exist considered if blood shortage exists. Although claret screening strategy has been improved, problems yet exist, particularly in many countries with loftier prevalence of blood-borne pathogen infections. While adopting NAT assays greatly shorten the window period that allows detection of many serologically undetectable infections possible, donations with MP-reactive but ID-non-reactive donations (MP+/ID−, divers as non-resolved donations) have been observed during daily screening process. These non-resolved donations may pose a significant risk for transfusion-transmitted infections in recipients, particularly in Communist china where HBV infection is endemic and anti-HBc testing cannot be implemented in routine claret screening. A Chinese multicenter study performed on 826,044 serologic negative donations in MPs of six identified that a full of 1,267 pools were reactive, of which 839 donations were reactive past ID-NAT, leaving 428(33.8%) not-resolved MPs (xix). These NAT initially reactive (IR)/not-resolved MPs might contain plasma from anti-HBc+ OBI donors with extremely depression and intermittently detectable HBV Deoxyribonucleic acid load but still potentially infectious (17, 19), raising urgent need for a practical algorithm to sort out which blood units can be safely transfused.
Previously, nosotros reported nearly half of the initial Ultrio plus NAT reactive, but further discriminatory exam negative donations were identified equally OBIs in ID-NAT screening setting (12), strongly suggesting a big proportion of samples had viral load below the 95% LOD of the Ultrio Plus ID-NAT assay (LOD: half-dozen.8 IU/donation) and also the MPX2.0 MP6 NAT assay (LOD: 13.8 IU/donation). Donations with such low viral loads had a high probability of being missed past the subsequent HBV ID-NAT, and this probability was adamant by Poisson distribution (twenty). The 5% LOD of the MPX assay is higher when testing in MP6 than in ID format (the 95% LOD of MPX2.0 test: two.3 IU/ml [ID format] and xiii.eight IU/ml [MP format]) (10), just a dilution factor of 6 is relatively small on the whole NAT detection endpoint probability bend that spans a concentration range of a factor of 100 between the 95 and the 5% LOD (19). Thus, information technology is likely to have a reactive effect in a puddle of six samples simply not in any of the individual samples. Some other possibility of MP-NAT reactive merely ID-NAT non-reactive (non-resolved) donations may be due to contamination, although this possibility is very low. To avoid contamination, we established a NAT laboratory with international standard, and all the screening procedures are fully automated, even for spiral capping. Furthermore, we adopted strict standard biosecurity and institutional safe procedures during the screening procedure.
In this study nosotros screened 17,226 pools (103,356 donations) by Roche MPX2.0 MP6-NAT, and nosotros identified 98 (0.57%, 95% CI 0.46–0.69%) MPs were initial reactive, among which 56/98 (57.i%) were resolved HBV Dna+. HBV DNA+ rate is 0.054% in this study cohort and is lower than screened by ID-NAT format reported in our previous study (12), likely considering of dilution cistron in MP-NAT and low-viral-load donations. Forty-ii of 17,226 (0.24%, 95% CI 0.18–0.33%) MPs were initially reactive, just all 6 donations from each MP were non-reactive (designated as non-resolved) when tested individually. This non-resolved percentage is in cyclopedia with the study in Commonwealth of australia (21), simply it is lower than the result from a national survey (19), probably due to the fact that the implementation of NAT in routine blood screening in Shenzhen Blood Center was ten years earlier than other Chinese blood centers. Afterward 42 non-resolved pools were further tested by Ultrio Plus ID-NAT, seven donations from 42 MPs (252 donations) were identified HBV DNA+ with anti-HBc+, and they became resolved. Since the probability of detection by NAT in low-viral-load samples follows a Poisson distribution, nosotros tested a total of 210 (0.xx%) donations in the remaining 35 not-resolved pools by 2.five ml large-volume extraction followed by nested PCR amplification and qPCR, and x donations from 35 MPs (210 donations) were farther identified HBV DNA+ with anti-HBc, indicating they were OBIs. It is undoubted that some low-viral-load donations were detected in MP6 format merely missed by ID-NATs. To sum upwards, 17 donations from 17/42 (40.4%) non-resolved pools (252 donations) containing HBV DNA+ were detected by additional alternative NATs, leaving 25/98 (25.5%) remaining non-resolved pools including 150 donations. These donations were released and transfused to recipients, posing potential threat to blood safety.
Previous studies estimated OBI transmission charge per unit for all components varied between 3 and 48% (8, 22, 23), which might be underestimated. A contempo mathematical model estimated that 3.3 and 14% of OBI donations undetected by NAT with LOD of iii.4 IU/ml might cause recipient infection by a blood component containing 20 ml and 200 ml of plasma, respectively (24). Infectious donations not detected by MP-NAT but reactive with ID-NAT take been reported (25, 26). Co-ordinate to clinical evidence and sequence identity, HBV transfusion manual in 9/31 recipients (29%) of claret components from donations undetected by the currently most sensitive NAT and HBsAg showed that even low levels of DNA in donors can exist infectious, and the revised lowest infectious dose was down to 0.14 IU/ml (7). Furthermore, two cases of transfusion-transmitted HBV infection were identified by donor-recipient sequence identity post-obit transfusion of xiv OBI donations missed by MP6 HBV DNA screening (8). In ID-NAT screening, NRR donations are non released for transfusion in most countries. Even so, for MP6 NAT, it is not an option to discard the units implicated in non-resolved pools. In this study, we used large-book viral nucleic acrid extraction, together with the highly sensitive nested PCR, to notice viral fragments, and we identified that 40% of non-resolved pools contained HBV DNA with anti-HBc. These donations could transmit HBV specially in immune compromised recipients. Our results indicated that even afterward MP-NAT screening in People's republic of china, donations with OBI nonetheless pose a potential residual threat for blood safety. Although transfusion-manual evidence is needed to substantiate this risk and anti-HBc and anti-HBs prevalence in recipients should as well be considered, our data nevertheless suggest that a proportion of non-resolved pools still comprise extremely low levels of HBV that may exist infectious, and more comprehensive resolving strategy should be considered for MP-NAT.
The presence of anti-HBs in add-on to anti-HBc indicates a resolved infection with persistent HBV DNA (27). In some countries such as Germany and Austria, claret units with anti-HBs levels greater than 100 IU/L are considered to be safe (28), and in Japan, anti-HBc-positive claret containing 200 IU/L or more of anti-HBs appears safe as a transfusion component (29). Notwithstanding, transmission of HBV from occult hepatitis B subjects occurred in the presence of concurrent neutralizing anti-HBs in the same specimen (xxx, 31). Data from organ transplantation also conspicuously proved that HBV Deoxyribonucleic acid in the presence of anti-HBs could be infectious in immunosuppressed patients (32). HBV DNA detected in some anti-HBs-positive samples in this report suggests that the absence of HBsAg and the presence of anti-HBs do not necessarily guarantee full safe. In the present report, 74.ii% donors were anti-HBs+ including 46% with anti-HBc and 28.ii% anti-HBs but, suggesting only a minor part of donors (28.2%) had a sure protection; well-nigh were induced by allowed response when non-vaccinated donor exposed to HBV or an anamnestic response to HBV when vaccinated donor exposed. 8 of 17 (47.1%) OBIs detected in not-resolved donations bear anti-HBs, suggesting that OBIs occur largely in individuals who have recovered from the infection but are unable to develop a totally effective immune control (33, 34). Anti-HBc alone has been observed either in a stage of late HBV immunity later on the decline of anti-HBs to undetectable levels or in the resolving phase of acute infection. Consistent with OBI, donated samples conveying anti-HBc alone are more infectious than those with low levels of anti-HBs (35). Even certain PCR negative "anti-HBc alone" individuals have been suspected to be potentially infectious (36). According to a contempo study, blood donors negative for both HBsAg and HBV DNA just reactive for anti-HBc might be HBV carriers with viral loads below the detection limit (37). In our present study, viii/17 non-resolved donations were identified equally OBIs with anti-HBc alone, and these OBIs may pose significant threat to blood safety.
It is well-known that anti-HBc is detectable during asymptomatic infections also equally throughout life after recovery from HBV infection with or without the presence of anti-HBs (38); therefore, anti-HBc is considered a key seromarker for OBI. Anti-HBc screening assays have the potential to exclude the bulk of OBIs undetectable by NAT (27, 39, 40), leaving only rare cases with escape mutants associated with the presence of anti-HBs alone (41). Many studies supported the utilize of serological markers such as anti-HBc to compensate less-sensitive NAT assays (42, 43). However, simply a relatively modest portion of OBIs can be identified by MP-NAT, which emphasizes the importance of anti-HBc testing and ID-NAT screening. In line with this, an American comprehensive study from 22.4 million blood donors screened by HBsAg, anti-HBc, and NAT revealed that but 43/404 (ten.six%) OBIs could be detected by MP-NAT, while most of OBIs (361/404, 89.four%) could but be identified by ID-NAT (44). These results indicated that the potential relative risk of OBI among MP- positive donations may exist small compared to that in MP-negative donations. Nevertheless, in countries such as in Cathay, where HBV endemic infections are high, anti-HBc screening may cause blood shortage. Considering the cost, ID-NAT is not a mandatory requirement for donor screening in Communist china; therefore, an alternative approach is to screen MP-positive donations with anti-HBc to identify those donations with potential OBIs to balance the price and safety. Ideally, blood centers should adopt ID-NAT screening especially in regions with loftier HBV endemicity, but in reality, unlike strategies have been used. We fabricated a crude assay of cost-effectiveness by comparing three strategies: (ane) HBsAg+ anti-HBc, (two) HBsAg+ MP6 NAT, (iii) HBsAg+ MP6+ anti-HBc in MP+/ID− (Tabular array 4). Undoubtedly, HBsAg and anti-HBc screening is the about highly price-effective, but it would overkill almost twoscore% anti-HBc+ donations, resulting in blood shortage. Calculation anti-HBc screening for MP+/ID− not-resolved donations would cost 5,004 RMB more only at to the lowest degree benefit from preventing the occurrence of vi transfusion-transmitted HBV cases (Table 4) with just deferring 0.11% anti-HBc+ donations to transfusion, merely obtaining 67.6 times benefit (46).

Tabular array 4 Toll-effectiveness analysis of 103,955 donors* in Chinese Shenzhen Claret Center.
It has been reported that mutations in S cistron or promoter and enhancer sequences of HBV genome could consequence in fake negative in HBsAg detection and NAT assays (47, 48). Although mutations from the consensus (wild type) may also be nowadays in a minority of not-OBI sequences, according to our sequence analysis, some OBIs indeed comport mutations in S gene or in BCP/PC, which most likely affect HBsAg detection (T126S, M133T, F134L, T143L, L175S) or inhibit HBV DNA replication (T1719G). The mutation bear on remains speculative without functional analysis, and we are in the process of setting upwards these assays to further confirm the biological significance of some of these mutations, especially in evasion of allowed surveillance and detection.
Although more than specific and sensitive NATs are urgently needed for donor screening in MP format, it is a long way to guarantee blood safe. Equally the majority of individuals with OBI accept very depression viral loads together with diverse mutations, the application of anti-HBc testing to evaluate not-resolved donors provides a better way to heighten blood safety in Prc. Additionally, some sensitive molecular methods similar nested PCR and existent-time PCR assay with loftier extraction book or nucleic acid hybridization for HBV DNA should be applied to identify OBIs in non-resolved blood donors.
Data Availability Statement
The original contributions presented in the report are included in the commodity/supplementary textile, further inquiries tin can be directed to the corresponding author.
Ethics Argument
This written report was reviewed and approved by the ideals committee of the Shenzhen Blood Middle. The written and informed consent form was obtained from each donor before donation.
Author Contributions
XY designed the experiments and wrote and reviewed the manuscript. LC reviewed, revised, and edited the manuscript. YZ, RL, TL, XZ, WX, JZ, MX participated in the written report design, performed the experiments, and collected and analyzed the information. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Nature and Science Fund of Shenzhen and Guangdong (JCYJ20190806112201646 and 2021A515010979) and Shenzhen Cardinal Medical Subject field Structure Fund (SZXK070) to XY and CAMS Initiative for Innovative Medicine (CAMS-2016-I2M-3-025 and CAMS-2017-I2M-B&R-fifteen), National Cardinal Research and Development Program (2018YFE0107500), Science and Engineering Partnership Program, Ministry of Science and Engineering of Red china (KY201904011) to LC.
Conflict of Involvement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed equally a potential conflict of interest.
Publisher's Note
All claims expressed in this commodity are solely those of the authors and practice non necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any production that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
HBV, Hepatitis B virus; HCV, hepatitis C virus; HIV, homo immunodeficiency virus; HBsAg, hepatitis B surface antigen; anti-HBc, hepatitis B core antigen; anti-HBs, hepatitis B surface antibody; HCC, hepatocellular carcinoma; NAT, nucleic acid testing; ELISA(s), enzyme-linked immuno-absorbent assay(s); ECLI, electrochemiluminescence immunoassay; ID, individual donation; LOD, limit of detection; MP(s), mini pool(s); ID, individual donation; NRR, non-repeat reactive; MP6, mini pools of six donations; OBI, occult hepatitis B virus infection; BCP/PC, basic cadre and pre-core promoter regions.
References
1. Raimondo R, Allain J-P, Brunetto MR, Buendia MA, Chen DS, Colombo Grand, et al. Statements From the Taormina Expert Meeting on Occult Hepatitis B Virus Infection. J Hepatol (2008) 49(four):652–7. doi: 10.1016/j.jhep.2008.07.014
PubMed Abstract | CrossRef Full Text | Google Scholar
2. Pollicino T, Squadrito G, Cerenzia G, Cacciola I, Raffa Thou, Craxi A, et al. Hepatitis B Virus Maintains its Prooncogenic Properties in the Case of Occult HBV Infection. Gastroenterology (2004) 126(i):102–10. doi: 10.1053/j.gastro.2003.x.048
PubMed Abstract | CrossRef Full Text | Google Scholar
3. Fopa D, Candotti D, Tagny CT, Doux C, Mbanya D, Tater EL, et al. Occult Hepatitis B Infection Among Blood Donors From Yaoundé, Republic of cameroon. Claret Transfus (2019) 17(6):403–8. doi: 10.2450/2019.0182-19
PubMed Abstract | CrossRef Full Text | Google Scholar
4. Candotti D, Lin CK, Belkhiri D, Sakuldamrongpanich T, Biswas S, Lin S, et al. Occult Hepatitis B Infection in Blood Donors From South East asia: Molecular Characterisation and Potential Mechanisms of Occurrence. Gut (2012) 61:1744–53. doi: 10.1136/gutjnl-2011-301281
PubMed Abstruse | CrossRef Total Text | Google Scholar
5. Huang CH, Yuan Q, Chen PJ, Zhang YL, Chen CR, Zheng QB, et al. Influence of Mutations in Hepatitis B Virus Surface Protein on Viral Antigenicity and Phenotype in Occult HBV Strains From Blood Donors. J Hepatol (2016) 57(2):720–9. doi: 10.1016/j.jhep.2012.05.009
CrossRef Full Text | Google Scholar
six. Yuen MF, Lee CK, Wong DK, Fung J, Hung I, Hsu A, et al. Prevalence of Occult Hepatitis B Infection in a Highly Endemic Surface area for Chronic Hepatitis B: A Study of a Big Blood Donor Population. Gut (2006) 59:1389–93. doi: ten.1136/gut.2010.209148
CrossRef Full Text | Google Scholar
7. Candotti D, Assennato SM, Laperche Southward, Allain J, Levicnik-Stezinar S. Multiple HBV Transfusion Transmissions From Undetected Occult Infections: Revising the Minimal Infectious Dose. Gut (2019) 68:313–21. doi: ten.1136/gutjnl-2018-316490
PubMed Abstract | CrossRef Full Text | Google Scholar
8. Spreafico M, Berzuini A, Foglieni B, Candotti D, Raffaele L, Guarnori I, et al. Poor Efficacy of Nucleic Acid Testing in Identifying Occult HBV Infection and Consequences for Safety of Claret Supply in Italian republic. J Hepatol (2015) 63(5):1068–76. doi: 10.1016/j.jhep.2015.06.016
PubMed Abstract | CrossRef Full Text | Google Scholar
9. Roth WK, Busch MP, Schuller A, Ismay S, Cheng A, Seed CR, et al. International Survey on NAT Testing of Claret Donations: Expanding Implementation and Yield From 1999 to 2009. Vox Sang (2012) 102(ane):82–ninety. doi: 10.1111/j.1423-0410.2011.01506.x
PubMed Abstruse | CrossRef Total Text | Google Scholar
x. Ohhashi Y, Pai A, Halait H, Ziermann R. Analytical and Clinical Performance Evaluation of the Cobas TaqScreen MPX Test for Use on the Cobas s 201 Arrangement. J Virol Methods (2010) 165(2):246–53. doi: ten.1016/j.jviromet.2010.02.004
PubMed Abstract | CrossRef Total Text | Google Scholar
11. Jarvis L, Becker J, Tender A, Cleland A, Queiros L, Aquiar A, et al. Evaluation of the Roche Cobas s 201 Arrangement and Cobas TaqScreen Multiplex Exam for Claret Screening: A European Multicenter Study. Transfusion (2008) 48(9):1853–61. doi: ten.1111/j.1537-2995.2008.01779.x
PubMed Abstract | CrossRef Total Text | Google Scholar
12. Ye Ten, Li T, Shao Due west, Zeng J, Hong W, Lu L, et al. Near Half of Ultrio Plus NAT Nondiscriminated Reactive Blood Donors Were Identified as Occult HBV Infection in South China. BMC Infect Dis (2019) nineteen:577–87. doi: 10.1186/s12879-019-4215-9
PubMed Abstract | CrossRef Full Text | Google Scholar
xiii. Enjalbert F, Krysztof DE, Candotti D, Allain JP, Stramer SL. Comparing of Seven Hepatitis B Virus (HBV) Nucleic Acid Testing Assays in Selected Samples With Discrepant HBV Markers Results From United States Blood Donors. Transfusion (2014) 54:2485–95. doi: 10.1111/trf.12653
PubMed Abstract | CrossRef Total Text | Google Scholar
14. Zheng 10, Ye X, Zhang L, Wang W, Shuai L, Wang A, et al. Characterization of Occult Hepatitis B Virus Infection From Blood Donors in People's republic of china. J Clin Microbiol (2011) 49:1730–seven. doi: 10.1128/JCM.00145-11
PubMed Abstract | CrossRef Full Text | Google Scholar
15. Yu D, Li 10, Monday V, Lu ZH, Liao XW, Han Y, et al. N-Glycosylation Mutations Inside Hepatitis B Virus Surface. J hepat (2014) 62(three):515–22. doi: 10.1016/j.jhep.2013.eleven.004
CrossRef Full Text | Google Scholar
xvi. Jiao F, Shen C, Ning J, Zhang T, Chen X, Lu F. HBV T1719G Mutation Reduced HBV Replication Through Mutant Enh 2 and Hbx Poly peptide in Vitro. J Viral Hepat (2019) 26(6):710–17. doi: ten.1111/jvh.13070
PubMed Abstract | CrossRef Full Text | Google Scholar
nineteen. Wang Fifty, Chang L, Xie Y, Huang C, Xu Fifty, Qian R, et al. What is The Meaning of a Nonresolved Viral Nucleic Acrid Test-Reactive Minipool? Transfusion (2015) 55(2):395–404. doi: 10.1111/trf.12818
PubMed Abstruse | CrossRef Full Text | Google Scholar
twenty. Weusten J, Vermeulen M, van Drimmelen H, Lelie N. Refinement of a Viral Manual Take a chance Model for Blood Donations in Seroconversion Window Stage Screened by Nucleic Acid Testing in Different Pool Sizes and Echo Test Algorithms. Transfusion (2011) 51(one):203–15. doi: x.1111/j.1537-2995.2010.02804.x
PubMed Abstract | CrossRef Total Text | Google Scholar
21. Margaritis AR, Brown SM, Seed CR, Kiely P, D'Agostino B, Keller AJ. Comparison of Two Automated Nucleic Acid Testing Systems for Simultaneous Detection of Human Immunodeficiency Virus and Hepatitis C Virus RNA and Hepatitis B Virus DNA. Transfusion (2007) 47(10):1783–93. doi: x.1111/j.1537-2995.2007.01343.x
PubMed Abstract | CrossRef Full Text | Google Scholar
22. Allain J, Mihaljevic I, Gonzalez-Fraile MI, Gubbe G, Holm-Harritshøj L, Garcia JM, et al. Infectivity of Claret Products From Donors With Occult Hepatitis B Virus Infection. Transfusion (2013) 53(vii):1405–15. doi: ten.1111/trf.12096
PubMed Abstract | CrossRef Full Text | Google Scholar
23. Kleinman SH, Lelie N, Busch MP. Infectivity of Man Immunodeficiency Virus-1, Hepatitis C Virus, and Hepatitis B Virus and Adventure of Transmission past Transfusion. Transfusion (2009) 49:2454–89. doi: 10.1111/j.1537-2995.2009.02322.x
PubMed Abstruse | CrossRef Full Text | Google Scholar
24. Weusten J, van Drimmelen H, Vermeulen Thousand, Lelie N. A Mathematical Model for Estimating Residuum Transmission Risk of Occult Hepatitis B Virus Infection With Different Blood Prophylactic Scenarios. Transfusion (2017) 57(3):841–ix. doi: 10.1111/trf.14050
PubMed Abstract | CrossRef Full Text | Google Scholar
25. Laperche S, Morel P, Deschaseaux M, Bouchardeau F, Alimardani One thousand, Guillaume N, et al. HIV Antibiotic Screening Remains Indispensable for Ensuring Viral Rubber of Claret Components Despite NAT Implementation. Transfusion (2013) 43(10):1428–32. doi: 10.1046/j.1537-2995.2003.00541.x
CrossRef Full Text | Google Scholar
26. Nübling CM, Heiden M, Chudy M, Kress J, Seitz R, Keller-Stanislawski B, et al. Experience of Mandatory Nucleic Acid Testing (NAT) Screening Across All Blood Organizations in Frg: NAT Yield Versus Breakthrough Transmissions. Transfusion (2009) 49(nine):1850–8. doi: 10.1111/j.1537-2995.2009.02212.x
PubMed Abstruse | CrossRef Total Text | Google Scholar
27. Taira R, Satake M, Momose S, Hino South, Suzuki Y, Murokawa H, et al. Residual Risk of Transfusion Transmitted Hepatitis B Virus (HBV) Infection Caused by Blood Components Derived From Donors With Occult HBV Infection in Japan. Transfusion (2013) 53(7):1393–404. doi: x.1111/j.1537-2995.2012.03909.x
PubMed Abstract | CrossRef Full Text | Google Scholar
29. Hoshi Y, Hasegawa T, Yamagishi N, Mizokami K, Sugiyama M, Matsubayashi G, et al. Optimal Titer of Anti-HBs in Blood Components Derived From Donors With Anti-HBc. Transfusion (2009) 59(8):2602–xi. doi: ten.1111/trf.15393
CrossRef Full Text | Google Scholar
30. Matsumoto C, Tadokoro K, Fujimura K, Hirakawa S, Mitsunaga S, Juji T. Analysis of HBV Infection After Claret Transfusion in Japan Through Investigation of a Comprehensive Donor Specimen Repository. Transfusion (2001) 41(3):878–84. doi: 10.1046/j.1537-2995.2001.41070878.x
PubMed Abstruse | CrossRef Full Text | Google Scholar
31. Levicnik-Stezinar Due south, Rahne-Potokar U, Candotti D, Lieli N, Allain JP. Anti-Hbs Positive Occult Hepatitis B Virus Carrier Blood Infectious in Two Transfusion Recipients. J Hepatol (2008) 48(five):1022–5. doi: ten.1016/j.jhep.2008.02.016
PubMed Abstract | CrossRef Full Text | Google Scholar
32. Castells L, Vargas V, Rodríguez-Frías F, Allende H, Jardí R, Margarit C, et al. Transmission of Hepatitis B by Transplantation of Livers From Donors Positive for Antibody to Hepatitis B Core Antigen. Transplant Proc (1999) 31(6):2464–five. doi: 10.1016/s0041-1345(99)00419-four
PubMed Abstruse | CrossRef Full Text | Google Scholar
33. Candotti D, Grabarczyk P, Ghiazza P, Roig R, Casamitjana N, Iudiconeb P, et al. Characterization of Occult Hepatitis B Virus Form Blood Donors Carrying Genotype A2 or Genotype D Strains. J Hepatol (2008) 49(4):537–47. doi: 10.1016/j.jhep.2008.04.017
PubMed Abstract | CrossRef Full Text | Google Scholar
36. Akcam FZ, Demir C. Acute Hepatitis B Infection Due to Erythrocyte Pause Obtained From 'Anti-HBc Alone' Positive Donor. Transf Med (2005) 15:61–2. doi: 10.1111/j.1365-3148.2005.00551.x
CrossRef Total Text | Google Scholar
37. Oluyinka OO, Tong HV, Bui Tien S, Fagbami AH, Adekanle O, Ojurongbe O, et al. Occult Hepatitis B Virus Infection in Nigerian Claret Donors and Hepatitis B Virus Transmission Risks. PloS One (2005) 10(1):131–9. doi: x.1371/journal.pone.0131912
CrossRef Full Text | Google Scholar
38. Urbani S, Fagnoni F, Missale M, Franchini M. The Function of Anti-Core Antibiotic Response in the Detection of Occult Hepatitis B Virus Infection. Clin Chem Lab Med (2010) 48(1):23–nine. doi: 10.1515/CCLM.2010.002
PubMed Abstruse | CrossRef Full Text | Google Scholar
39. Vermeulen M, Coleman C, Walker Eastward, Koppleman M, Lelie N, Reddy R. Transmission of Occult HBV Infection past ID-NAT Screened Blood. Vox Sang (2014) 107(Suppl one):146–eight. doi: 10.1111/trf.12218
CrossRef Full Text | Google Scholar
twoscore. Seed CR, Maloney R, Kiely P, Bell B, Keller A, Pink J, et al. Infectivity of Blood Components From Donors With Occult Hepatitis B Infection – Results From an Australian Look Back Programme. Voice Sang (2015) 108(1):113–22. doi: x.1111/vox.12198
PubMed Abstract | CrossRef Total Text | Google Scholar
41. Stramer SL, Wend U, Candotti D, Foster GA, Hollinger FB, Dodd RY, et al. Nucleic Acrid Testing to Detect HBV Infection in Blood Donors. N Engl J Med (2011) 364(iii):236–47. doi: x.1056/NEJMoa1007644
PubMed Abstruse | CrossRef Total Text | Google Scholar
42. Hourfar MK, Walch LA, Geusendam G, Dengler T, Janetzko K, Gubbe K, et al. Sensitivity and Specificity of Anti-HBc Screening Assays – Which Analysis Is Best for Blood Donor Screening? Int J Lab Hematol (2009) 31:649–56. doi: 10.1111/j.1751-553X.2008.01092.ten
PubMed Abstract | CrossRef Full Text | Google Scholar
44. Dodd RY, Nguyen ML, Krysztof DE, Notari EP, Stramer SL. Blood Donor Testing for Hepatitis B Virus in the U.s.: Is There a Case for Continuation of Hepatitis B Surface Antigen Detection? Transfusion (2018) 58:2166–70. doi: 10.1111/trf.14784
PubMed Abstract | CrossRef Full Text | Google Scholar
45. Ye Ten, Yang B, Zhu W, Zheng X, Du P, Zeng J, et al. Six-Year Airplane pilot Written report on Nucleic Acid Testing for Blood Donations in Red china. Trans Aphe Sci (2013) 49(ii):318–22. doi: 10.1016/j.transci.2013.08.005
CrossRef Full Text | Google Scholar
46. Cun W, He Q, Zhang J, Xiang Z. Cost Benefit Analysis of 43 714 Donors Blood Nucleic Acid Testing Practical to Screening in Wuhan, China. Chin J Blood Transfusion (2014) 27:166–8. doi: 10.13303/j.cjbt.issn.1004-549x.2014.02.0019
CrossRef Full Text | Google Scholar
47. Hass M, Hannoun C, Kalinina T, Sommer Grand, Manegold C, Gunther Due south. Functional Analysis of Hepatitis B Virus Reactivating in Hepatitis B Surface Antigen-Negative Individuals. Hepatology (2005) 42(1):93–103. doi: 10.1002/hep.20748
PubMed Abstract | CrossRef Total Text | Google Scholar
48. Allain JP. International Collaborative Written report Proposal for the Label of Occult Hepatitis B Virus Infection Identified by Nucleic Acrid or Anti-Hbc Screening. Vocalization Sang (2007) 92(3):254–7. doi: ten.1111/j.1423-0410.2006.00874.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Source: https://www.frontiersin.org/articles/10.3389/fimmu.2021.699217/full
0 Response to "Hepatitis B Virus Dna Detectable Again"
Post a Comment